Chapter 3: Summarizing data
Contents
Chapter 3: Summarizing data#
import pandas as pd
import sidetable
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import norm
import rpy2.robjects as ro
from rpy2.robjects.packages import importr
from rpy2.robjects import pandas2ri
pandas2ri.activate()
from rpy2.robjects.conversion import localconverter
# import NHANES package
base = importr('NHANES')
with localconverter(ro.default_converter + pandas2ri.converter):
NHANES = ro.conversion.rpy2py(ro.r['NHANES'])
NHANES = NHANES.drop_duplicates(subset='ID')
Table 3.1#
pd.DataFrame(NHANES.PhysActive.value_counts(dropna=False))
| PhysActive | |
|---|---|
| Yes | 2972 |
| No | 2473 |
| NaN | 1334 |
Table 3.2#
table_df = NHANES.stb.freq(['PhysActive']).drop(
['cumulative_count', 'cumulative_percent'], axis=1)
table_df['RelativeFrequency'] = table_df.percent / 100
table_df = table_df.rename(columns={
'count': 'AbsoluteFrequency',
'percent': 'Percentage'})
table_df.style.hide(axis="index")
| PhysActive | AbsoluteFrequency | Percentage | RelativeFrequency |
|---|---|---|---|
| Yes | 2972 | 54.582185 | 0.545822 |
| No | 2473 | 45.417815 | 0.454178 |
Table 3.3#
NHANES_sleep = NHANES.query('SleepHrsNight > 0')
table_df = NHANES_sleep.stb.freq(
['SleepHrsNight'])
table_df['RelativeFrequency'] = table_df.percent / 100
table_df = table_df.rename(columns={
'count': 'AbsoluteFrequency',
'percent': 'Percentage'}).sort_values(by='SleepHrsNight').drop(
['cumulative_count', 'cumulative_percent'], axis=1)
table_df.style.hide(axis="index")
| SleepHrsNight | AbsoluteFrequency | Percentage | RelativeFrequency |
|---|---|---|---|
| 2 | 9 | 0.178749 | 0.001787 |
| 3 | 49 | 0.973188 | 0.009732 |
| 4 | 200 | 3.972195 | 0.039722 |
| 5 | 406 | 8.063555 | 0.080636 |
| 6 | 1172 | 23.277061 | 0.232771 |
| 7 | 1394 | 27.686197 | 0.276862 |
| 8 | 1405 | 27.904667 | 0.279047 |
| 9 | 271 | 5.382324 | 0.053823 |
| 10 | 97 | 1.926514 | 0.019265 |
| 11 | 15 | 0.297915 | 0.002979 |
| 12 | 17 | 0.337637 | 0.003376 |
Figure 3.2#
fig, ax = plt.subplots(1, 2, figsize=(12,8))
sns.histplot(NHANES_sleep.SleepHrsNight, ax=ax[0], discrete=True)
sns.histplot(NHANES_sleep.SleepHrsNight, ax=ax[1], stat='density', discrete=True)
<Axes: xlabel='SleepHrsNight', ylabel='Density'>
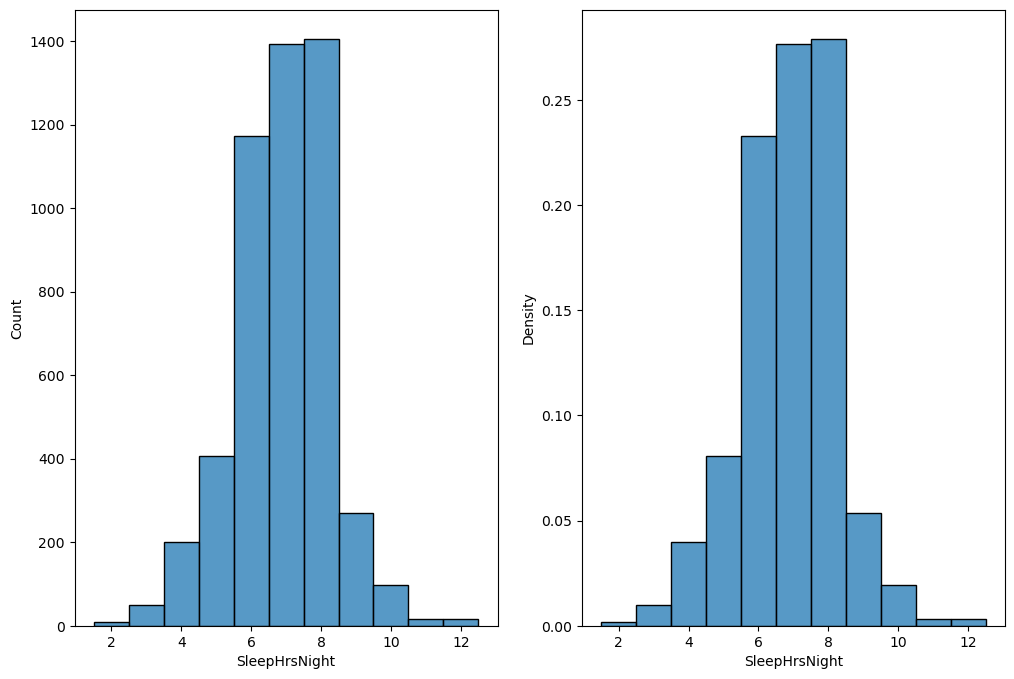
Figure 3.3#
fig, ax = plt.subplots(1, 2, figsize=(12,8))
sns.histplot(NHANES_sleep.SleepHrsNight, ax=ax[0],
element="step", fill=False, cumulative=True, )
sns.histplot(NHANES_sleep.SleepHrsNight, ax=ax[0], discrete=True)
sns.histplot(NHANES_sleep.SleepHrsNight, ax=ax[1], stat='density',
element="step", fill=False, cumulative=True, )
sns.histplot(NHANES_sleep.SleepHrsNight, ax=ax[1], stat='density', discrete=True)
<Axes: xlabel='SleepHrsNight', ylabel='Density'>

Figure 3.4#
fig, ax = plt.subplots(1, 2, figsize=(12,8))
sns.histplot(NHANES.Age, binwidth=1, ax=ax[0])
sns.histplot(NHANES.Height, binwidth=1, ax=ax[1])
<Axes: xlabel='Height', ylabel='Count'>

Figure 3.5#
fig, ax = plt.subplots(2, 2, figsize=(12,8))
NHANES['isChild'] = NHANES.Age < 18
NHANES_adult = NHANES.dropna(subset=['Age', 'Height']).query('Age > 17')
sns.histplot(NHANES, x='Height', hue='isChild', binwidth=1, ax=ax[0][0])
ax[0][0].set_title('A. All individuals')
sns.histplot(NHANES_adult, x='Height', binwidth=1, ax=ax[0][1])
ax[0][0].set_title('B. Adults only')
sns.histplot(NHANES_adult, x='Height', binwidth=.1, ax=ax[1][0])
ax[1][0].set_title('B. Adults only (bin width=.1)')
Text(0.5, 1.0, 'B. Adults only (bin width=.1)')
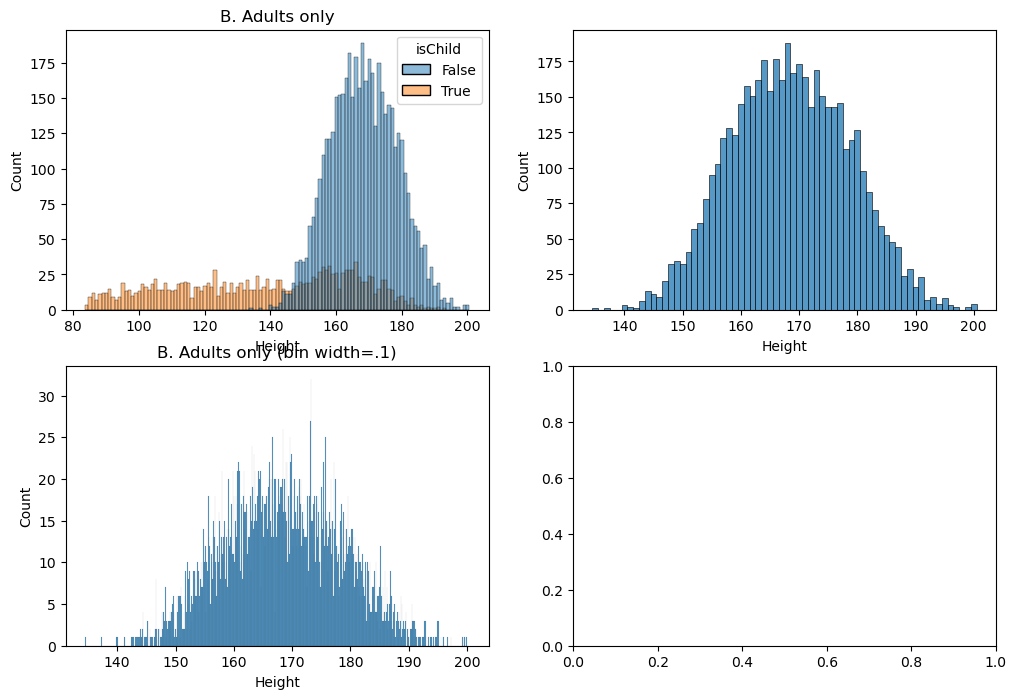
Table 3.5#
NHANES_adult['Height'][44:50]
96 169.6
97 169.8
101 167.5
102 155.2
104 173.8
107 174.5
Name: Height, dtype: float64
Figure 3.6#
# first update the summary to include the mean and standard deviation of each
# dataset
# NOTE: some NA values get turned into large negative numbers in the conversion from R, so remove those
pulsedata = NHANES_adult.dropna(subset=['Pulse']).query('Pulse > 0')[['Pulse']]
heightdata = NHANES_adult.dropna(subset=['Height']).query('Height > 0')[['Height']]
pulse_summary = pulsedata.describe()
pulse_summary.loc['binwidth'] = np.diff(np.histogram_bin_edges(pulsedata.Pulse, 'fd'))[0]
height_summary = heightdata.describe()
height_summary.loc['binwidth'] = np.diff(np.histogram_bin_edges(heightdata.Height, 'fd'))[0]
heightDist = pd.DataFrame({'x': np.arange(height_summary.loc['min'][0], height_summary.loc['max'][0], .1)})
heightDist['norm'] = norm.pdf(
heightDist.x, height_summary.loc['mean'][0], height_summary.loc['std'])
pulseDist = pd.DataFrame({'x': np.arange(pulse_summary.loc['min'][0], pulse_summary.loc['max'][0], .1)})
pulseDist['norm'] = norm.pdf(
pulseDist.x, pulse_summary.loc['mean'][0], pulse_summary.loc['std'])
fig, ax = plt.subplots(1, 2, figsize=(12,8))
sns.histplot(heightdata.Height, stat='density', binwidth=height_summary.loc['binwidth'][0], color='gray', ax=ax[0])
ax[0].plot(heightDist.x, heightDist.norm, lw=2, color='b')
sns.histplot(pulsedata.Pulse, stat='density', binwidth=pulse_summary.loc['binwidth'][0], color='gray', ax=ax[1])
ax[1].plot(pulseDist.x, pulseDist.norm, lw=2, color='b')
[<matplotlib.lines.Line2D at 0x7f56a7c1d930>]
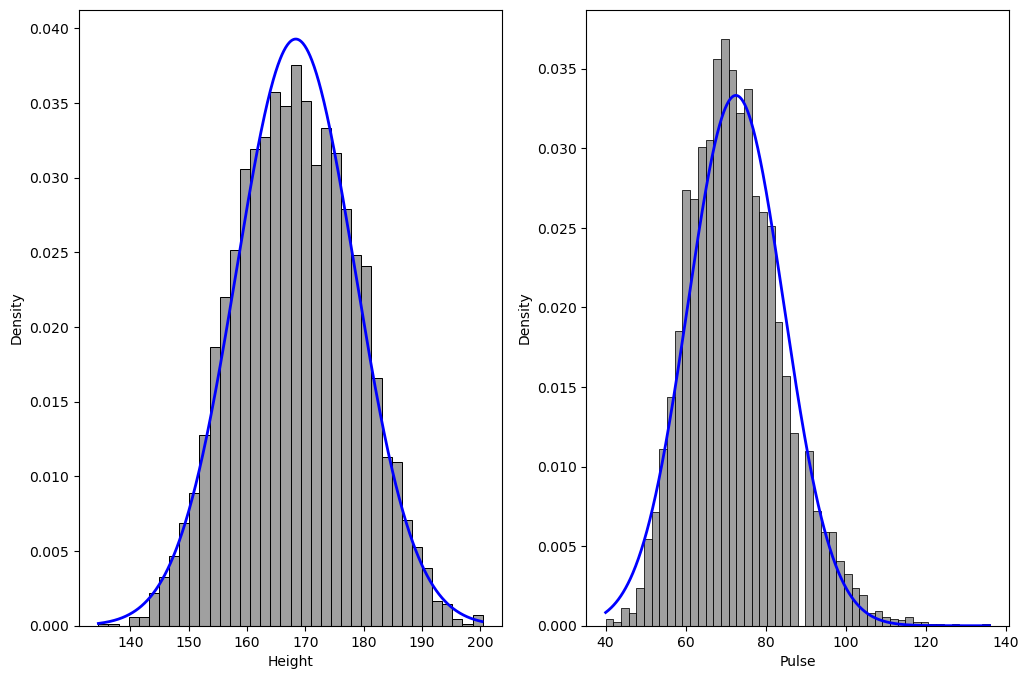
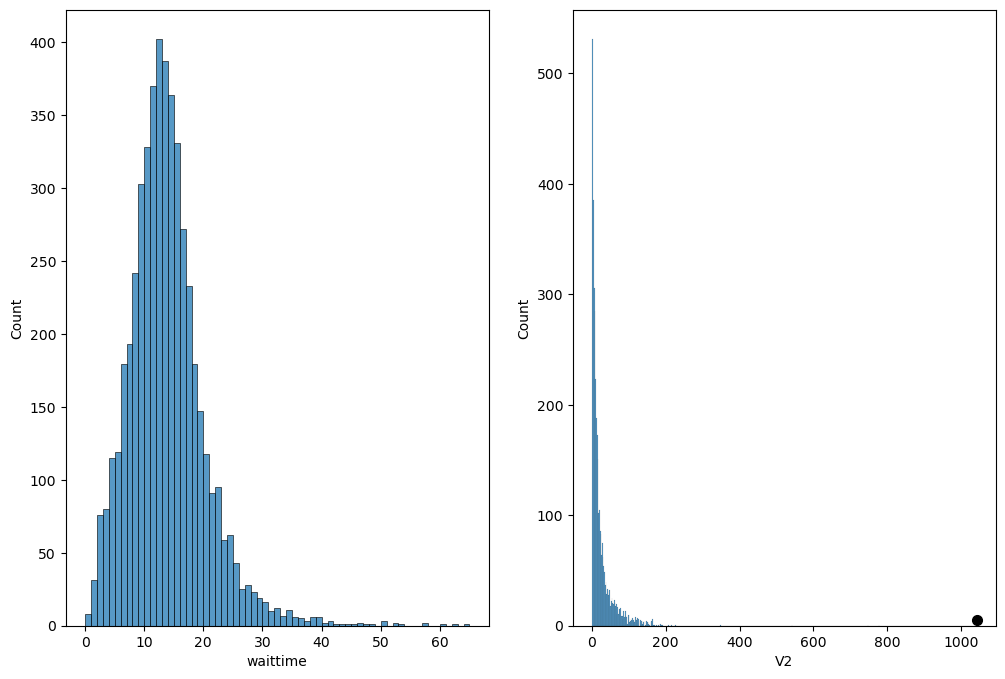
Figure 3.7#
waittimes = pd.read_csv('https://raw.githubusercontent.com/statsthinking21/statsthinking21-figures-data/main/04/sfo_wait_times_2017.csv')
fig, ax = plt.subplots(1, 2, figsize=(12,8))
sns.histplot(waittimes.waittime, binwidth=1, ax=ax[0])
fbdata = pd.read_csv('https://raw.githubusercontent.com/statsthinking21/statsthinking21-figures-data/main/04/facebook_combined.txt', names=['V1', 'V2'], delimiter=' ')
friends_table = fbdata.groupby('V1').count()
sns.histplot(friends_table.V2, binwidth=2, ax=ax[1])
plt.plot([friends_table.max()['V2']], [5], marker='.', markersize=14, color='k')
[<matplotlib.lines.Line2D at 0x7f56a6236e00>]